Problem
N6-methyladenosine (m6A) is the most prevalent modification found on eukaryotic mRNA. This modification is implicated in the development of cancer, stem cell differentiation, control of circadian rhythm, and more. m6A bases cannot be detected directly by sequencing, because the m6A modification does not change the base pairing properties and cannot be detected by reverse transcription. MeRIP is a commonly used method to detect m6A modifications in mRNA. This method involves immunoprecipitation followed by sequencing of the enriched sample; methylated regions are detected as peaks in transcript coverage from immunoprecipitated RNA compared to input RNA. This method is time-consuming and can suffer from variability in detection of methylation. Overall, detection of N6-methyladenosine down to the single-nucleotide level has been impeded by a lack of robust analytical methods.
Solution
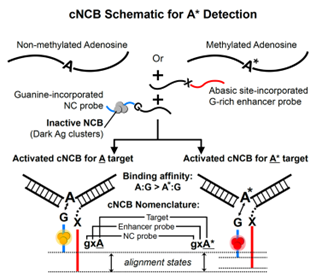
The Yeh Lab at The University of Texas at Austin has developed chameleon NanoCluster Beacons (cNCBs), capable of detecting methylated adenosine at single-base resolution. The system uses a three-way junction hybridization configuration with a target. When the probe binds to a target, silver clusters interact with the enhancer sequence and are activated. At the junction, an abasic site on the enhancer probe is a non-interacting, neutral site, while guanine on the probe is a key base that interacts with either N6-methyladenosine or adenine on the target. Interactions between adenine and guanine are expected to be stronger than that of N6-methyladenosine and guanine, leading to the ability to detect the N6-methyladenosine sites by differences in the alignment.
Value proposition
- Dr. Yeh’s technology allows for discrimination between N6-methyladenosine and adenine, identifying N6-methyladenosine sites at single-nucleotide resolution.
- The detection method is low in cost, easy to prepare, and has high fluorescence target-to-background ratios, making detection robust and consistent.
- The technology is enzyme-free and highly reproducible.
About the inventor
Dr. Hsin-Chih Yeh is an Associate Professor of Biomedical Engineering at the Cockrell School of Engineering. He is a leading expert in nanobiosensor development, cancer biomarker detection, single-molecule spectroscopy for biomolecule and nanomaterial characterization, 3D molecular tracking, and super-resolution imaging.

